Abstract
Background and aims: Deletions in the long arm of chromosome 9, del(9q), are recurrent cytogenetic aberrations in myeloid neoplasms. While relatively common in patients with acute myeloid leukemia (AML), del(9q) occurs less frequently in other myeloid malignancies such as myelodysplastic syndromes (MDS) or myeloproliferative neoplasms (MPN). To present, cases with del(9q) in MDS and MPN are poorly characterized on a molecular level. We set out to define the size and minimally deleted region (MDR) of del(9q) as well as the spectrum of accompanying gene mutations in patients with MDS or MPN. Furthermore, the prognostic impact of del(9q) in MDS is still under debate. According to the revised international prognostic scoring system (IPSS-R), MDS patients with isolated del(9q) fall into the cytogenetic intermediate risk group (median overall survival, OS, of 32 months) based on the presence of a single aberration (Greenberg et al, Blood 2012). We therefore aim to evaluate if this risk stratification is appropriate for our patient cohort.
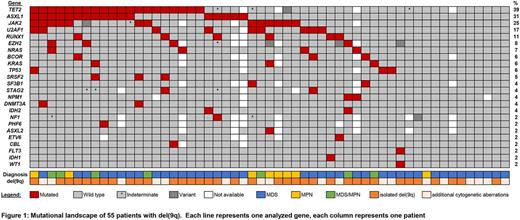
Patients and methods: A total of 55 (26 female and 29 male) patients with MDS (n=40), MPN (n=8) and MDS/MPN overlap (n=7) were included in this study. The median age was 75 years (range: 23-94 years). Conventional cytogenetic analysis was performed for all patients. Cases (n=38) with sufficient clone size were investigated by array CGH (SurePrint G3 ISCA CGH+SNP Microarray, Agilent, Waldbronn, Germany). Mutation status was examined using an amplicon-based NGS panel targeting 40 genes commonly affected in myeloid neoplasia.
Results: In 36/55 (65%) of patients, del(9q) was the sole cytogenetic aberration. Additional recurrent cytogenetic abnormalities included unbalanced rearrangements, n=8; del(5q), n=5; del(11q), del(20q), +8, +21 (n=2, each). Clinical follow-up data was available for 16 MDS patients with isolated del(9q), median OS was 35 months (range, 1- 91 months). In cases with array CGH data, size of the 9q deletion varied between 2.2 and 70.5 Mb (median, 33.9 Mb). We identified a MDR of 3.6 Mb ranging from genomic position 85,716,785 to 89,340,559 (UCSC hg19). A total of 21 genes are located in the MDR, including FRMD3, UBQLN1, KIF27, HNRNPK, SLC28A3 and NTRK2, which have previously been described in the MDR of del(9q) AML patients. Driver gene mutations were identified in 43/55 (78%) of patients. The number of mutated genes ranged from 1-5 (median: n=2). We did not observe significant differences in the number of mutated genes between younger or older patients (cut-off: 70 years), and patients with isolated del(9q) or additional cytogenetic aberrations. However, patients without evidence for a gene mutation (n=12) were significantly younger than patients with respective mutations (median age 70 vs. 76 years, p=0.014). Recurrently mutated genes included TET2 (39%), ASXL1 (31%), JAK2 (25%), U2AF1 (17%), RUNX1 (11%), EZH2 (8%), NRAS (7%), BCOR, KRAS, TP53 (6% each) and SRSF2 (5%). ASXL1 and TET2 were frequently co-mutated, as 12/20 (60%) of TET2 mutated cases also harbored ASXL1 mutations (p< 0.001). Overall, we identified a high prevalence of TET2 mutations in patients with del(9q). This association was even more pronounced when we limited our analyses to patients with isolated del(9q), where we observed a TET2 mutation frequency of 45%. In 7/31 (23%) of patients without TET2 mutation, other genes associated with epigenetic regulation, namely DNMT3A, ASXL1, EZH2, WT1, IDH1 and IDH2, were affected, underlining a high mutation rate (49% of total cohort) in this functional class. Mutations linked to the spliceosome machinery, SF3B1, SRSF2 and U2AF1, were mutated in 13/55 (24%) of patients in the total cohort and in 9/40 (23%) patients with MDS.
Conclusions: Deletions in 9q are rare aberrations in MDS and MPN. Our analyses provide novel insights into the molecular profile of this cytogenetic subgroup. 1) Our patient cohort with MDS or MPN and del(9q) demonstrated a minimal deleted region in concordance with previous reports on the MDR of del(9q) AML patients, indicating a general pathogenic mechanism. 2) We identified gene mutations in 78% of our patient cohort, TET2 and other epigenetic regulators were mutated in 49% of patients with del(9q), suggesting cooperative pathogenic potential. 3) We observed a median OS of 35 months in patients with MDS and isolated del(9q), confirming the current stratification according to IPSS-R.
Hartmann: MLL Munich Leukemia Laboratory: Employment. Haferlach: MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Meggendorfer: MLL Munich Leukemia Laboratory: Employment. Fasan: MLL Munich Leukemia Laboratory: Employment. Stengel: MLL Munich Leukemia Laboratory: Employment. Zenger: MLL Munich Leukemia Laboratory: Employment. Zieschang: MLL Munich Leukemia Laboratory: Employment. Kern: MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Haferlach: MLL Munich Leukemia Laboratory: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal